Publications
Single-molecule tracking to determine the abundances and stoichiometries of freely-diffusing protein complexes in living cells: past applications and future prospects
J. Prindle, O. de Cuba, A. Gahlmann
Journal of Chemical Physics, 2023, 159, 071002
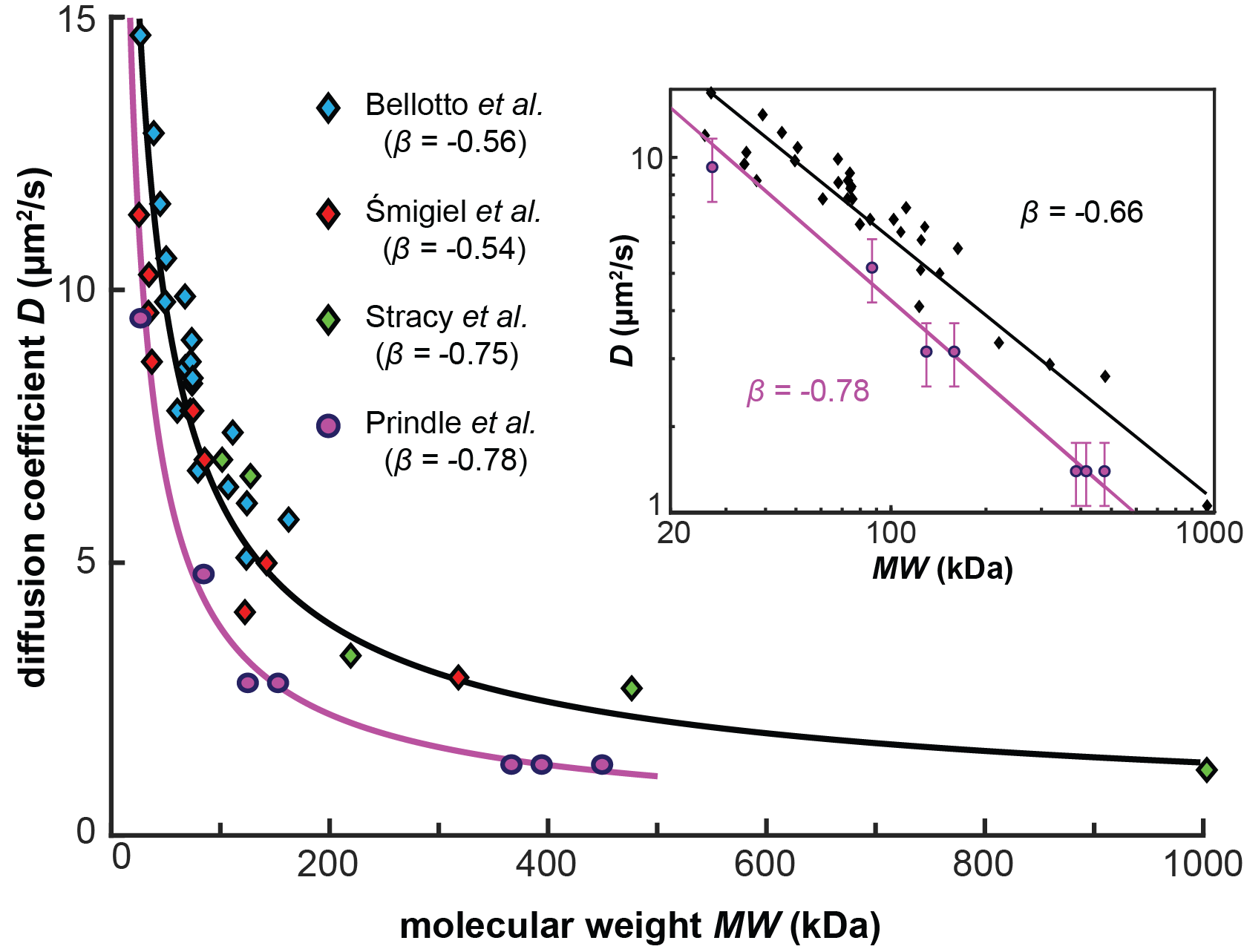
Distinct complexes containing the cytosolic type III secretion system ATPase resolved by 3D single-molecule tracking in live Yersinia enterocolitica.
J. Prindle, J. Rocha, Y. Wang, A. Diepold, A. Gahlmann
Microbiology Spectrum, 2022, 10, 6, e01744-01722
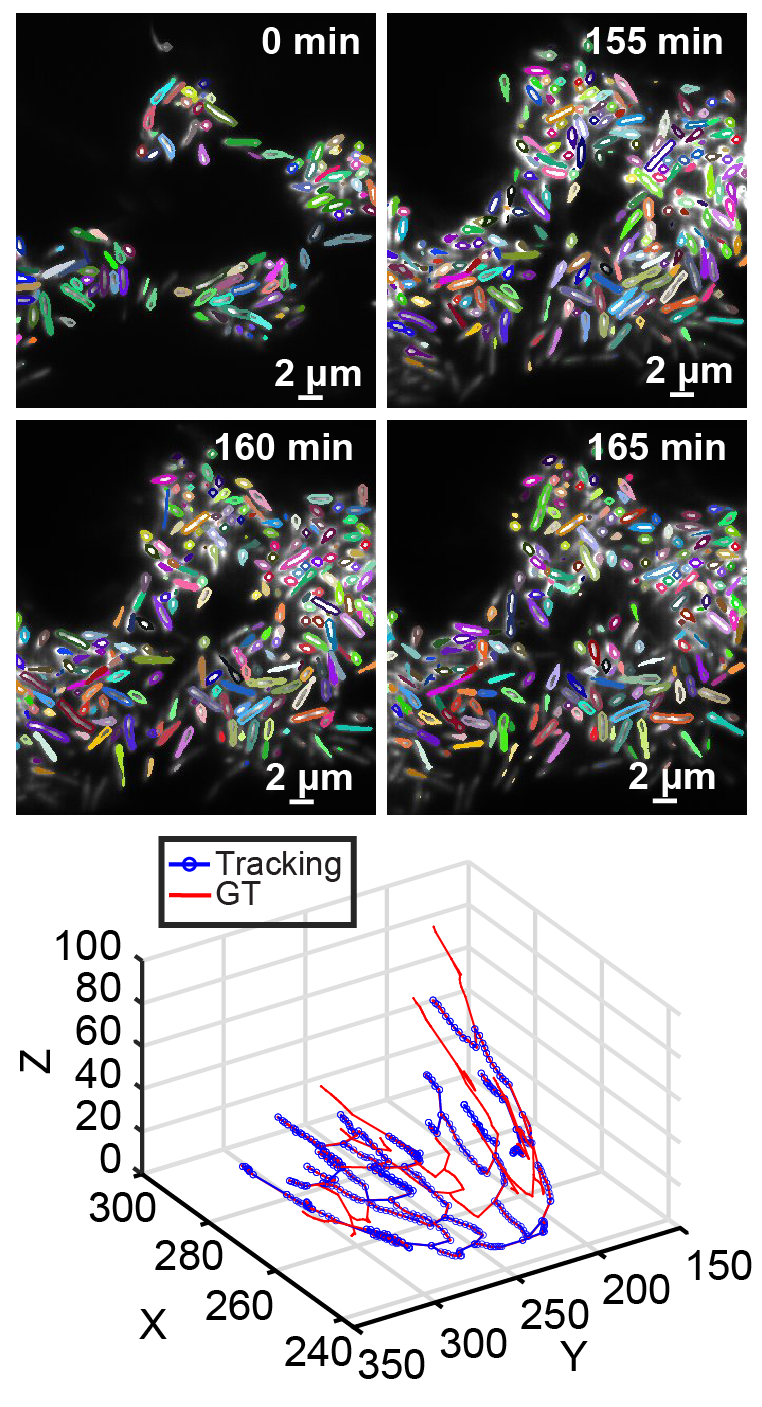
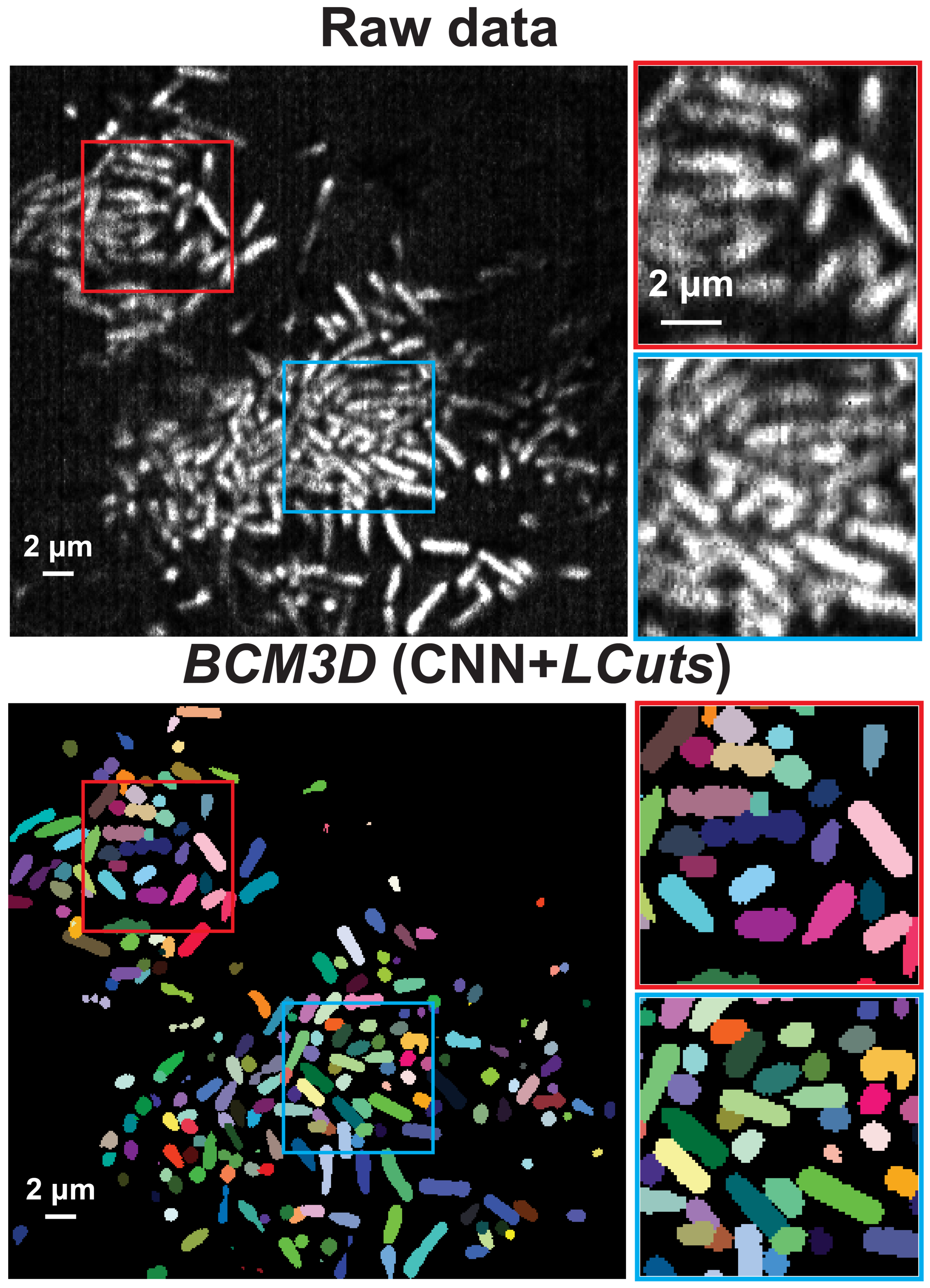
BCM3D 2.0: Accurate Segmentation of Single Bacterial Cells in Dense Biofilms using Computationally Generated Intermediate Image Representations
J. Zhang, Y. Wang, E. D. Donarski, T. T. Toma, M. T. Miles, S. T. Acton, A. Gahlmann
npj Biofilms and Microbiomes, 2022, 8, 99, 1-13
See also the associated blog post.
BCM3D 2.0 is publicly available on GitHub (https://github.com/GahlmannLab/BCM3D-2.0) and through the Open Science Framework (https://osf.io/m4637/).
Realistic-Shape Bacterial Biofilm Simulator for Deep Learning-Based 3D Single-Cell Segmentation
T.T. Toma, Y. Wu, J. Wang, A. Srivastava, A. Gahlmann, S. T. Acton
IEEE International Conference on Image Processing (ICIP) 1-5 (2022)
Optically Accessible Microfluidic Flow Channels for Noninvasive High-Resolution Biofilm Imaging Using Lattice Light Sheet Microscopy
J. Zhang, M. Zhang, Y. Wang, E. Donarski, A. Gahlmann
J. Phys. Chem B, 2021, 125 (44), 12187-12196
Graph-Theoretic Post-Processing of Segmentation with Application to Dense Biofilms
J. Wang, M. Zhang, J. Zhang, Y. Wang, A. Gahlmann, S.T. Acton
IEEE Transactions on Image Processing, 2021, 30, 8580-8594
Non-Invasive Single-Cell Morphometry in Living Bacterial Biofilms
M. Zhang, J. Zhang, Y. Wang, J. Wang, A.M. Achimovich, S.T. Acton, A. Gahlmann
Nature Communications, 2020, 11, 6151
Facile Synthesis and Metabolic Incorporation of m-DAP Bioisosteres Into Cell Walls of Live Bacteria
A. Apostolos, J. Nelson, J.R. Silva, J. Lameira, A.M. Achimovich, A. Gahlmann, C. Alves, M. Pires.
ACS Chemical Biology, 2020, 15, 2966-2975
Structural Similarity Image Analysis for Detection of Adenosine and Dopamine in Fast-Scan Cyclic Voltammetry Color Plots
P. Puthongkham, J. Rocha, J.R. Borgus, M. Ganesana, Y. Wang, Y. Chang, A. Gahlmann, B.J. Venton.
Analytical chemistry, 2020, 92 (15), 10485-10494.
Reprogramming fatty acyl specificity of lipid kinases via C1 domain engineering
T.B. Ware, C.E. Franks, M.E. Granade, M. Zhang, K.-B. Kim, K.-S. Park, A. Gahlmann,
T.E. Harris, and K.-L. Hsu. Nature Chemical Biology 2020, 16, 170-178.
Enabling technologies in super-resolution fluorescence microscopy: Reporters, labeling, and methods of measurement
A.-M. Achimovich, H. Ai, A. Gahlmann. Current Opinion in Structural Biology , 2019, 58, 224-232
LCuts: Linear Clustering of Bacteria using Recursive Graph Cuts
J. Wang, T. Batabyal, M. Zhang, J. Zhang, A. Aziz, A. Gahlmann, S.T. Acton
2019 IEEE International Conference on Image Processing (ICIP) 1575-1579 (2019)
Resolving cytosolic diffusive states in bacteria by single-molecule tracking
J. Rocha, J. Corbitt, T. Yan, C. Richardson, A. Gahlmann. Biophysical Journal, 2019, 116, 1970-1983
Computational correction of spatially-variant optical aberrations in 3D single-molecule localization microscopy
T. Yan, C. Richardson, M. Zhang, A. Gahlmann. Optics Express, 2019, 27, 12582
Single-Molecule Tracking Microscopy - A Tool For Determining The Diffusive States Of Cytosolic Molecules
J. Rocha, A. Gahlmann. The Journal of Visualized Experiments (JoVE), 2019, 151, e59387
Single-molecule tracking in live Yersinia enterocolitica reveals distinct cytosolic complexes of injectisome subunits
J. Rocha, C. Richardson, M. Zhang, C. Darch, E. Cai, A. Diepold, A. Gahlmann
Integrative Biology, 2018, 10, 502 (Cover Article)
BACT-3D: A level set segmentation approach for dense multi-layered 3D bacterial biofilms
J. Wang, R. Sarkar, A. Aziz, A. Vaccari, A. Gahlmann, S. Acton
2017 IEEE International Conference on Image Processing (ICIP) 330-334 (2017)
Prior to UVA
Bacterial Scaffold Directs Pole-Specific Centromere Segregation
J.L. Ptacin, A. Gahlmann, G.R. Bowman , A.M. Perez, A.R.S. von Diezmann, M.R. Eckart, W.E. Moerner, and
L. Shapiro. Proc. Natl. Acad. Sci. USA, 2014, 111, E2046
Exploring Bacterial Cell Biology with Single-Molecule Tracking and Super-Resolution Imaging
A. Gahlmann and W.E. Moerner. Nat. Rev. Microbiol., 2013, 12, 9 (Cover Article)
Quantitative Multicolor Subdiffraction Imaging of Bacterial Protein Ultrastructures in Three Dimensions
A. Gahlmann, J.L. Ptacin, G. Grover, S. Quirin, A.R.S. von Diezmann, M.K. Lee, M.P. Backlund, L. Shapiro, R. Piestun, and W.E. Moerner. Nano Lett., 2013, 13, 987
Structural Dynamics of Free Amino Acids in Diffraction
I-R. Lee, A. Gahlmann, and A.H. Zewail, Angew. Chem. Int. Ed., 2012, 51, 99
Direct Structural Determination of Conformations of Photoswitchable Molecules by Laser Desorption-Electron Diffraction
A. Gahlmann, I-R. Lee, and A.H. Zewail. Angew. Chem. Int. Ed., 2010, 49, 6524
Structure of Isolated Biomolecules by Electron Diffraction-Laser Desorption: Uracil and Guanine
A. Gahlmann, S.T. Park, and A.H. Zewail. J. Amer. Chem. Soc., 2009, 131, 2806 (Cover Article)
Ultrafast Electron Diffraction Reveals Dark Structures of the Biological Chromophore Indole
S.T. Park, A. Gahlmann, Y. He, J.S. Feenstra, and A.H. Zewail, Angew. Chem. Int. Ed., 2006, 47, 9496
Ultrashort Electron Pulses for Diffraction, Crystallography and Microscopy: Theoretical and Experimental Resolutions
A. Gahlmann, S.T. Park, and A.H. Zewail. Phys. Chem. Chem. Phys., 2008, 10, 2894
Ultrafast Electron Diffraction: Structural Dynamics of Molecular Rearrangement in the NO Release from Nitrobenzene
Y. He, A. Gahlmann, J.S. Feenstra, S.T. Park, and A.H. Zewail, Chemistry - An Asian Journal, 2006, 1, 56
Investigating the Effect of the Zwitterion/Lactone Equilibrium of Rhodamine B on the Cybotactic Region of the Acetonitrile/scCO2 Cosolvent
A. Gahlmann, K.D. Kester, and S.G. Mayer, J. Phys. Chem. A., 2005, 109, 1753